Deciphering cell fate decision at single-cell resolution
Grün Lab
Substantial variability of mRNA levels across cells of the same type has been observed in any organism studied, ranging from bacteria and yeast to mammals. Gene expression variability can have diverse origins. Heterogeneity of cell states following extrinsic or intrinsic stimuli, e. g., due to cell cycle, or changes in the microenvironment, can be one reason for cell-to-cell transcriptome differences. Moreover, many genes are transcribed in bursts rather than at a constant rate, leading to substantially different transcript numbers in individual cells of the same type, commonly addressed as gene expression noise. Gene expression variability causes fluctuations in protein levels, and can entail physiological differences between cells.
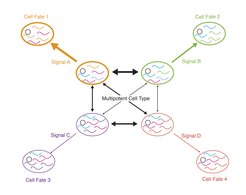
In our lab we investigate how stem cells robustly maintain their multipotent state and reliably execute differentiation programs with spatial and temporal precision in the presence of gene expression noise. We propose a model in which stem cells can exist in different metastable states, which are primed towards distinct lineages by subtle transcriptome modulations (Figure 1). While transitions between these stages occur in the multipotent state, reinforcement of a primed state by random fluctuations of lineage determining factors or signalling events from the microenvironment lead to commitment towards a terminal fate. We explore the molecular mechanisms underpinning the transition from a plastic multipotent state towards a lineage-restricted committed state. We have a particular focus on the integration of external cell fate determinants, i.e., signals from the microenvironment, with cell-intrinsic driving forces of fate bias such as gene expression noise.
