
Bioinformatics Facility
The regulation of gene expression is controlled by multiple mechanisms, such as the sequence-specific binding of transcription factors to DNA, epigenetic signals and a dynamic chromatin state. An unbiased understanding of these processes requires access to large-scale experiments and the capacity to analyze genome-wide data. In close interaction with the Deep Sequencing Facility of the MPI-IE, we streamline the primary analysis and quality control of data, which emerges at an unprecedented rate and resolution from a wide range of new sequencing experiments.
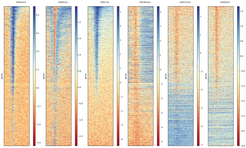
Heatmaps are useful tools to visualize genome-wide data, such as important histone marks around all human genes.
In the context of scientific collaborations we are also developing new methods and tools for quality control, analysis and visualization of deep-sequencing data (e.g. deepTools2, HiCExplorer, snakePipes). Together with the Deep-Sequencing Facility, we actively contribute to larger networks of epigenomic research (CRC992 Medical Epigenetics) and support these projects with our expertise in deep-sequencing data generation and analysis.
The Bioinformatics Group is operating a powerful Data Center (more than 1000 cores and 1 PetaByte storage) for the primary analysis of sequencing data and other large-scale biocomputing. For our internal users we host extensive web services, workflows and customized tools that help with data management, visualizations, standardized analyses and data sharing.
To help our colleagues with the interpretation of genome-wide data, the group offers regular training courses and interactive tutorials.
Selected Publications
Find more publications of Thomas Manke and his team on PubMed.
