Differentiation of immune cells
Grün Lab
As a model of multi-lineage differentiation we are studying cells of the hematopoietic system with a focus on lymphocytes. Years of intense research have revealed the major hematopoietic cell types as well as multipotent progenitor populations with the help of flow cytometry on the basis of a relatively small number of cell surface markers. The development of sensitive high-throughput single-cell sequencing has revolutionized the identification of cell types and differentiation trajectories, and recent studies are changing our model of hematopoietic differentiation (Figure 1).
We are studying differentiation of murine hematopoietic cell populations in primary lymphoid organs, e.g., bone marrow and fetal liver, as well as differentiation of tissue-resident cells of the innate immune system (Herman et al., Nature Methods, 2018; Sagar et al., The EMBO Journal, 2020; Zeis et al., Immunity, 2020). Our aim is to elucidate the dependence of hematopoietic cell fate decision and lineage output on the local microenvironment, in order to explain differences in hematopoiesis across tissues, as well as temporal heterogeneity of immune cell differentiation during life. In more detail, we are aiming to identify tissue-specific progenitor states and regulatory mechanisms underpinning local lineage choice, and to understand the role of gene expression variability in this context.
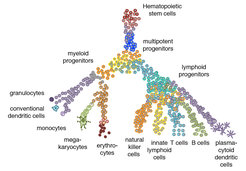
By means of single-cell sequencing analysis we are creating high-resolution map of cell states and the differentiation trajectories connecting these states. These maps are used to generate hypotheses on progenitor states and mechanisms of fate commitment, which are functionally analysed with the help of mouse models and in vitro culture systems. To obtain information on the interactions of hematopoietic cells with neighbouring cells in the local microenvironment, we are integrating single-cell sequencing methods with spatial imaging-based analysis using highly multiplexed single-molecule FISH in tissue sections.
To facilitate translation of our research into biomedically relevant therapeutic approaches, we are combining the analysis of mouse models and human patient material.
