
Sample Preparation
Deep Sequencing Facility
Prior sequence analysis any sample must be prepared. Key is to preserve biological information during the complete preparation workflow.
The deep sequencing facility supports sample preparation for multiple assays most common are Transcriptome-, Chromatin-, Whole Genome-, or Methylome Assays. Input material and requirements vary per assay. For each assay “standard”-, “low”- or even “single-cell”- preparation technologies are available.
Furthermore, preparation assays differ between short- and long-read sequencing technologies. For short-read sequencing (Illumina) nucleic acids are during preparation artificially fragmented. Long-read sequencing protocols aim to preserve the length of the nucleic acids.
The following section gives an overview of the main preparation techniques in use. Please be aware that a huge number of different preparation workflows are existing. Still, most of the published techniques are to some extend modifications of the main technologies used in the field. Especially downstream of every workflow (sequencing library preparation) concepts and techniques in use are quite redundant.
Preparation workflows depend on the RNA fraction of interest.
For genexpression analysis poly-A containing messenger RNA (mRNA) molecules are purified by usage of poly-T oligos bound to magnetic beads. To analyze coding and non-coding RNA molecules, ribosomal RNA (rRNA) is depleted using target specific oligos.
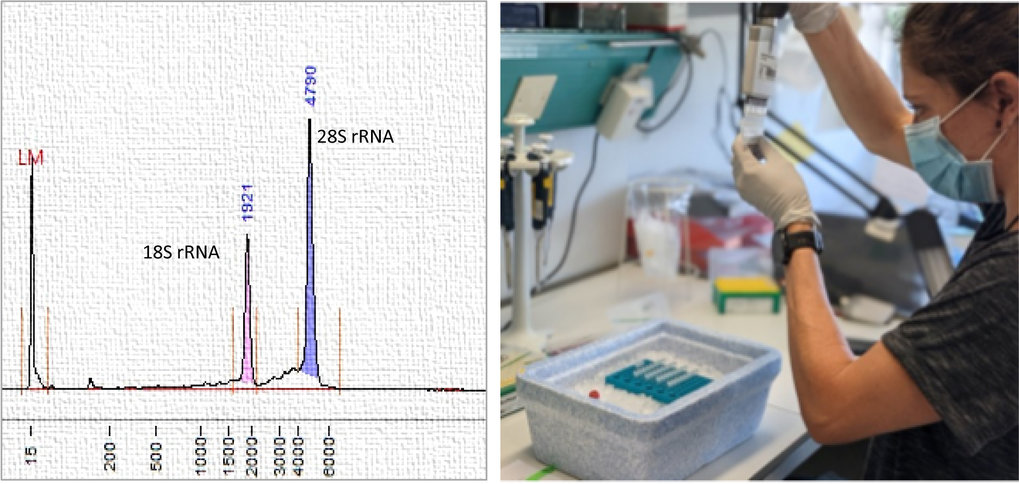
Left: Analysis of high-quality RNA. Right: Preparation of RNA samples for sequencing.
After capture of polyadenylated mRNAs or depletion of rRNA background full-length double-stranded cDNA is prepared. For short-read sequencing (Illumina) cDNA is fragmented, adaptor ligated and PCR-amplified. For long-read sequencing (Oxford Nanopore) intact cDNA is adapter-ligated and (if needed) PCR-amplified. For both sequencing technologies (long-, short-read) multiple indexing options are available to optimize usage of sequencing capacities or refine sequence accuracy (usage of unique molecular identifier, UMIs).
For small-RNA sequencing (micro RNAs (miRNAs), small non-coding RNAs), adapters are ligated to each end of the RNA molecule, reverse transcribed and PCR-amplified. A gel purification step (BluePippin) is necessary to clear libraries from byproducts.
Typically Protein/DNA interactions (ChIP-Seq, CUT&RUN), open chromatin regions (ATAC-Seq) or Chromatin conformation (HiC) are analyzed.
For Chromatin immunoprecipitation (ChIP-Seq) crosslinking preserves Protein/DNA interactions. Upon chromatin isolation, target specific antibodies are used to select regions of interest. Subsequently DNA fragments are released and during library preparation adapter ligated and PCR amplified.
CUT&RUN (Cleavage Under Targets & Release Using Nuclease) works on native Chromatin and utilizes target-specific primary antibodies and pAG-MNase (fusion of Proteins A and G to Micrococcal Nuclease) to isolate protein-DNA complexes for analysis using next generation sequencing.
ATAC-Seq is a technique used to mark transposase-accessible chromatin (profile open chromatin regions). In ATAC-Seq, genomic DNA is exposed to Tn5, a highly active transposase. Tn5 simultaneously fragments DNA and adds sequencing primers (a process known as tagmentation).
HiC is used to study chromosome interactions. Technically: Crosslinking of chromatin is followed by restriction enzyme digestion and biotinylation of overhangs prior ligation of nearby ends. Biotinylated ligation junctions are purified and DNA released for library preparation.
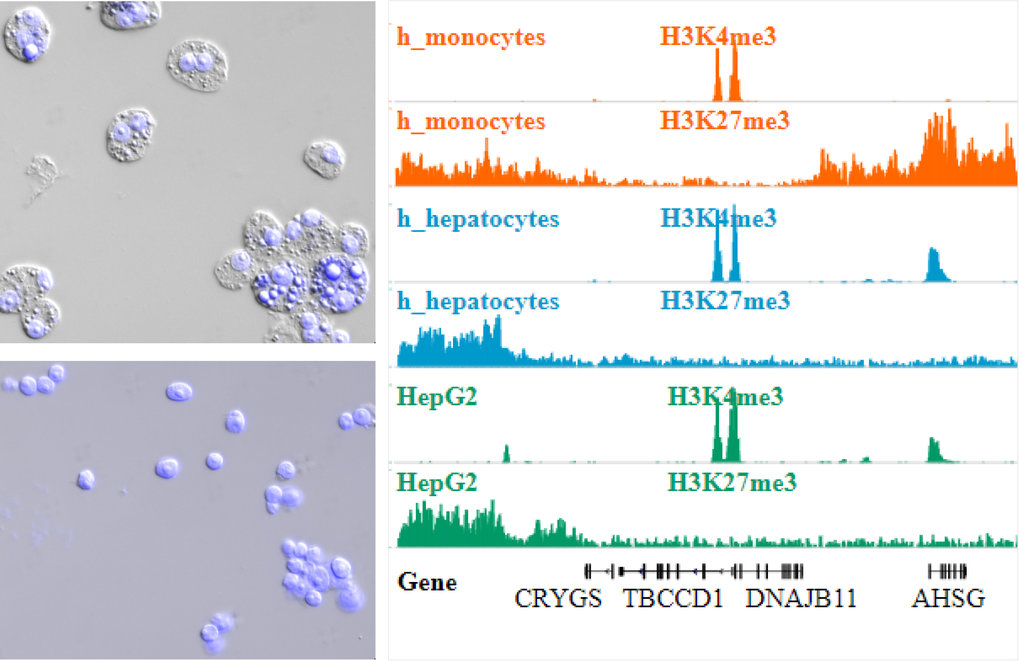
ChIP-Seq including Nuclei Extraction by Sonication (NEXSON) for Chromatin preparation. Top Left: HepG2 cells before nuclei extraction. Bottom left: HepG2 cells after nuclei extraction. (Pictures in DAPI (blue, nuclei) and differential interference contrast (DIC) channels were taken before and after ultrasound treatment ). Right: Read coverage profiles of histone modification ChIP-seq for human monocytes (orange), hepatocytes (blue) and HepG2 cell line (green) prepared by NEXSON.
WGS is a comprehensive method to analyze the entire genome. The technique is rather simple. High-molecular-weight genomic DNA (HMW gDNA) is isolated from material of interest. For short-read sequencing DNA fragments are fragmented using ultrasound (< 1kb) and sequencing adapters ligated. Assays in use are sensitive enough to omit PCR amplification prior sequencing.
If Oxford Nanopore Technology (ONT) is used for long-read genome sequencing, read lengths is limited only by the molecule lengths in the sample. Therefore, preserving intactness of DNA prior sequencing adapter ligation and sequencing is key. Depending on the DNA extraction method ONT data can be considered “standard long read” (10–100kb) and “ultra-long read” (greater than 100kb).
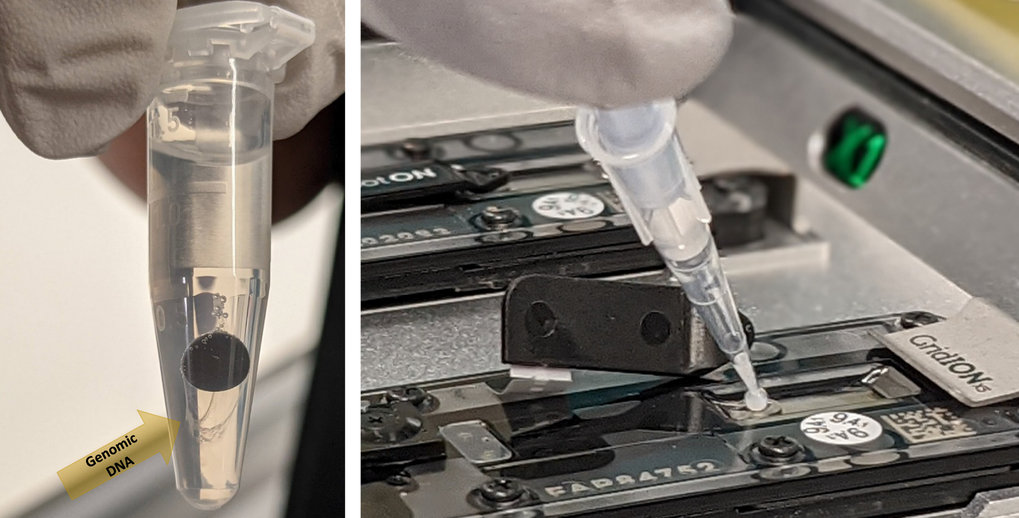
Left: DNA extraction using Nanobind magnetic discs (Circulomics). Allows for extraction of high molecular weight (50 – 300 kb) and ultra high molecular weight (50 kb – 1Mb) DNA. Right: Loading high molecular weight DNA on a GridION flowcell for long read sequencing (Oxford nanopore technology).
Methyl-Seq is used to detect pattern of DNA methylation. Bisulfite treatment is gold standard for methylome analysis. Treatment with bisulfite converts cytosine residues to uracil but leaves 5-methylcytosine (5-mC) unaffected. Hence bisulfite treated DNA retains only methylated cytosines.
Using Oxford Nanopore Technology (ONT) epigenetic modifications such as methylation pattern can be directly detected in DNA and RNA molecules, making this technology extremely valuable.
For single cell assays the “10X Genomics” technology is applied. Innovative barcoding concepts are facilitating Single Cell Gene Expression, Single Cell ATAC and Single Cell Multiome (Geneexpression + ATAC from the same cell) analysis. The 10X technology relies on partitioning of thousands of cells into nanoliter-scale Gel Beads-in-emulsion (GEMs). All generated cDNA molecules share a common 10x Barcode. Dual Indexed libraries are generated and sequenced and 10x Barcodes are used to associate individual reads back to the individual partitions.

10X Genomics Technology for single cell analysis. Left: Chromium Controller (10X Genomics), Right: GEMs (Gel bead-in-emulsion, microscopy picture.)