Using an organelle-system approach to reveal the identity of metabolic circuits
Rambold Lab
Project
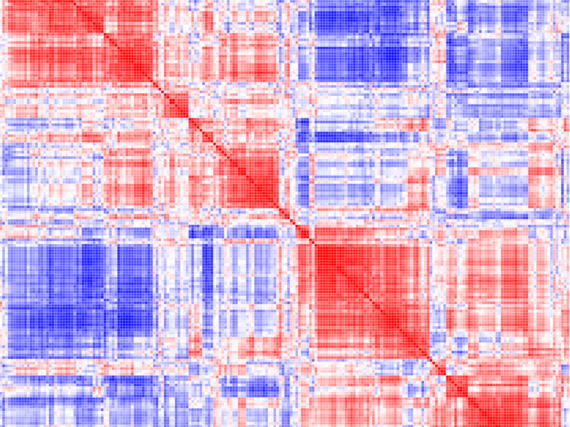
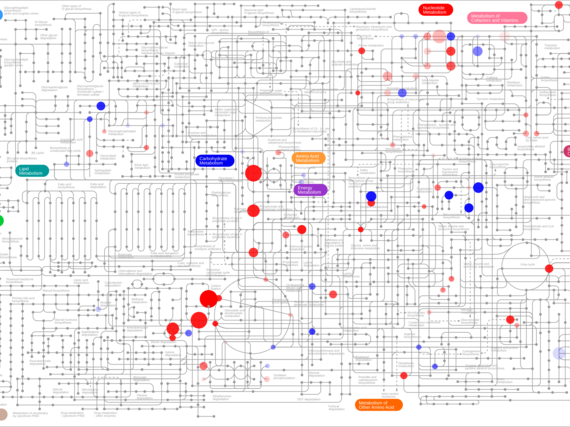
Based on mostly pair-wise organelle studies from yeast and non-immune cells, it is becoming clear that many, potentially all, organelles are able to communicate with one another via direct- or indirect interaction modes. In this project we aim at gaining a comprehensive understanding about large-scale organelle communication networks (via direct and indirect communication modes) and delineate how they are used to adapt organelle homeostasis and metabolism in healthy and diseased immune cells.
We are using targeted and systems approaches, combining high-resolution multicolor imaging with gene expression, proteomic and metabolic profiling on the population and single cell/organelle level to gain an integrated view of organelle-networks in primary macrophages, during inflammation, infection and resolution processes. By comparing our results with macrophage responses from models of human organelle-linked immune disorders, we aim at untangling primary defects from secondary spreading effect along the organelle networks. These may provide novel intervention points in the effort to ameliorate immune cell defects in affected patients.