MSR repeat RNA organize a heterochromatic RNA-nucleosome scaffold
Lab Jenuwein
Another hallmark of heterochromatic regions is the occurrence of non-coding RNA, which originate from the repeat sequences. Attenuation and subsequent silencing of the initial repeat-derived transcription appears crucial for heterochromatin formation, as the absence or the excess of transcriptional activity fails to establish and maintain heterochromatic marks. Repeat-derived non-coding RNA could therefore facilitate recruitment of chromatin factors by serving as guide RNA and/or by constituting a structural component of a distinct heterochromatin configuration.
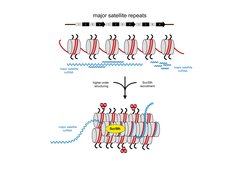
MSR transcripts largely lack poly(A) signals and remain chromatin associated. The data reveal RNA:DNA hybrids and single-stranded RNA to be important for the recruitment of the Suv39h KMT and suggest that a higher-order RNA-nucleosome scaffold is the physiological template for the stable association of Suv39h enzymes to chromatin (Velazquez Camacho et al. 2017) (Figure 1). We are now extending these studies to examine secondary structures of MSR transcripts that allow the formation of RNA:DNA hybrids or fold into distinct stem loops for the possible RNA interaction of chromatin factors.
We will also investigate 5meC and me6A RNA methylation of repeat-derived transcripts and modulate the expression of endogenous MSR repeats by targeting of TALEN and/or dCas9 directed transcriptional activators and transcriptional repressors to heterochromatin.
