Function of neuron-specific 3’ UTRs
Hilgers Lab
Neuron-specific ultra-long 3’UTRs also occur in humans; their function is unknown.
We think that a balanced expression of ultra-long 3’UTRs limits phenotypes of neurodegeneration in disease and in normal aging. Our findings indicate that deleting the neuron-specific part of mRNA sequences can cause serious neurological phenotypes in vivo.
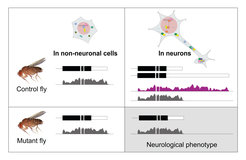
Experimental strategy to determine the function of individual ultra-long 3’UTRs. Control flies express the short mRNA isoform in most cells, and the long mRNA isoform in neurons. Mutant flies lack the neuronal part of the 3’UTR and display neurological phenotypes. mRNA isoforms are depicted schematically with the corresponding RNA-seq tracks.
© Valérie Hilgers
We aim to establish the functional impact of ultra-long 3’UTRs on neuronal function. Approaches include functional genetics, behavior analysis, single-molecule imaging, proteomics and transcriptomics.
