Research
Lab Akhtar
MOF, MOF-associated complexes and H4K16ac in flies
Males have one X chromosome and females two in both flies and mammals. However, the requirement for most of the protein products encoded by the X chromosome is invariant across the sexes: in other words, females do not produce double the X-linked proteins that males do. This therefore poses a dosage problem: how do the different “doses” of chromosomes translate into the same “dose” of proteins? This biological problem is solved by the epigenetic process of dosage compensation. In mammals one of the two X chromosomes in females is shut down through the concerted action of multiple repressive epigenetic regulators. In flies, expression from the single male X chromosome is instead upregulated through chromosome-wide H4K16 hyperacetylation. The MSL complex contains the histone acetyltransferase MOF responsible for depositing H4K16ac throughout the X chromosome. Our lab has made multiple seminal discoveries into the dosage compensation process in flies, which serves as a model for regulation of gene dosage on a chromosome-wide level.
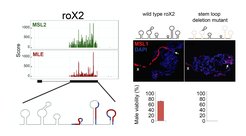
Left, upper: MSL2 and MLE iCLIP profiles in the Drosophila melanogaster Cl8 male cell line.
Left, lower: stem loop organization of roX2 RNA based on SHAPE analysis
Right: Deletion of multiple roX2 stem loops abolishes the X-chromosomal targeting of MSL1 and leads to lethality in males. Expression of wildtype and stem loop deleted roX2 (both under the control of UAS) was driven ectopically in a roX1/roX2 null background using the tub-GAL4 driver.
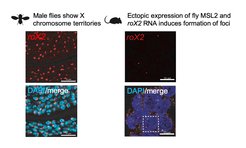
In addition to five protein components, the fly MSL complex contains two long noncoding RNAs: roX1 and roX2. Our work has uncovered that these lncRNAs have defined modular stem loop structures which are remodeled by the RNA helicase component MLE before incorporation into the MSL complex (Ilik et al 2013 Molecular Cell; Quinn et al 2014 Nature Biotechnology). Our most recent work has revealed that the interaction between the C-terminal domain of MSL2 and the nascent roX RNAs found in proximity of the X chromosome produces a gel-like state which drives MSL complex specificity for this chromosome (Keller Valsecchi et al 2021 Nature).Although MSL complex binding to the X chromosome is initially nucleated at special genomic locations known as high-affinity sites (HAS), the complex subsequently coats the entire chromosome through the process of “spreading”. By performing Hi-C analyses we have found that the distinct 3-dimensional chromosome conformation of the fly X chromosome facilitates the spread of the roX long noncoding RNAs. The sites at which the MSL complex first engages with roX RNAs (the high-affinity sites; HAS) are preferentially found near topologically associating domain (TAD) boundaries and show enhanced long-range contacts (Ramírez et al 2015 Molecular Cell).
DHX9 is the mammalian orthologue of the RNA helicase MLE. However, DHX9 does not interact with the MSL proteins and is therefore not believed to be a part of the MSL complex in mammals. We have shown that DHX9 is required for post-transcriptional RNA homeostasis and specifically for genome defense against transcripts of endogenous Alu elements. DHX9 binds and uses its RNA helicase activity to resolve the inverted repeats of Alu retrotransposable elements (Aktaş et al 2017 Nature).
Structural work into the MSL complex has produced insights into the molecular architecture of these proteins. Our crystal structures of mouse MSL1 with MOF and MSL3 as well as of MSL1 with MSL2 have proven invaluable in pinpointing essential amino acids at interaction interfaces within the MSL complex (Hallacli et al 2012 Molecular Cell; Kadlec et al 2011 Nature Structural & Molecular Biology). Interestingly both MSL1 and KANSL1 engage MOF via their PEHE domains, and MOF’s interaction with one or the other is mutually exclusive. Thus, MSL1 and NSL1 PEHE domains enable MOF’s “choice” between the two complexes. The crystal structures additionally delineated the interaction interface between MSL3 and MSL1, which plays a crucial role in modulating the enzymatic activity of MOF.
Although MOF and H4K16ac have historically been studied for their roles in dosage compensation, recent evidence suggests that MOF additionally has sex-independent functions in flies as well. Already in 2008 our lab showed that, in addition to coating the X chromosome in males, MOF binding is found at the promoters of a significant proportion of autosomal genes. We found that this autosomal binding is also observed in females (Kind et al 2008 Cell). More recently, the lab has shown that the H4K16ac mark is intergenerationally maintained from egg to embryo, where it is required for the appropriate activation of genes important for embryonic development in both male and female flies (Samata et al 2020 Cell).
MOF, MOF-associated complexes and H4K16ac in mammals
The mammalian MSL complex does not display selectivity for the X chromosome. Instead, mammalian MSL2 appears to regulate expression of individual genes rather than whole chromosomes (Chelmicki et al 2014 eLife; Keller Valsecchi et al 2018 Nature Communications). The MSL complex influences the expression of Tsix, the major repressor of Xist, thereby contributing to regulation of X-inactivation (mammalian dosage compensation) (Chelmicki et al 2014 eLife). Mutations in the gene encoding MSL complex component MSL3 in humans lead to the MSL3/Basilicata-Akhtar syndrome, suggesting that MSL3 has important developmental functions (Basilicata et al 2018 Nature Genetics). All the MSL3 syndrome patient mutations identified to date are located in the portion of the gene sequence encoding the MRG domain, which our crystal structure had revealed as the interface between MSL3 and MSL1 (Kadlec et al 2011 Nature Structural & Molecular Biology) and known to be required to stimulate the enzymatic activity of the MOF enzyme towards histones.
MOF has been shown to have important roles in tissue development and homeostasis in mice. Since full-body knockout of Mof results in embryonic lethality (Thomas et al 2008 Molecular Cell Biology; Gupta et al 2008 Molecular Cell Biology), we rely on conditional systems to delete Mof specifically in tissues of interest. This approach has permitted us to delineate the function of MOF in the blood lineage, heart, kidney and brain.

MOF participates in lineage specification in the hematopoietic system in mice. MOF guides chromatin accessibility dynamics along erythropoiesis, and is specifically involved in priming the chromatin state of hematopoietic stem cells for transcriptional engagement of the erythroid program (Pessoa Rodrigues et al 2020 Science Advances). Mice with Mof deletion in hematopoietic stem cells exhibit altered genomewide H4K16 acetylation, changed chromatin accessibility and ultimately develop anemia.
Mice with cardiac- and skeletal-muscle-specific deletion of Mof die of heart failure between postnatal days 16-23. Examination of Mof-knockout hearts revealed signs of hypertrophy and progressive accumulation of interstitial fibroblasts (Chatterjee et al. 2016 Cell). This extensive fibrosis may indicate that MOF plays a role in cardiomyocyte regeneration.
Mof knockout in kidney podocytes has a surprisingly mild phenotype until the animals encounter acute stress in the form of Adriamycin (Skeikh et al 2015 Oncogene). This work uncovered that MOF in podocytes is dispensable at steady state, but takes on an essential role in mediating the transcriptional response when the kidney encounters acute stress or injury.
In 2006 the lab discovered that, in addition to the MSL complex, MOF also associates with another distinct set of proteins collectively known as the NSL complex (Mendjan et al 2006 Molecular Cell). Four of the NSL complex components (NSL1, NSL2, NSL3 and MBD-R2) are found exclusively in the NSL complex, whereas the other three can also be members of other chromatin complexes: MOF also associates with the MSL complex, MCRS1 can also assemble into the INO80 complex, and WDR5 is part of the MLL/COMPASS complexes. There had previously been suggestions in the literature that the NSL and MLL/COMPASS complexes interact via their shared WDR5 subunit to form a supercomplex. However, crystal structures from our lab have shown that the WDR5 interactions with NSL and MLL/COMPASS complexes are mutually exclusive (Dias et al 2014 Genes & Development).
The NSL complex is a major transcriptional regulator in flies (Raja et al 2010 Molecular Cell; Lam et al 2012 PLoS Genetics). All components of this complex are also conserved in mammals and are involved in regulating pluripotency in embryonic stem cells (ESCs) (Chelmicki et al 2014 eLife). Furthermore, MOF, MSL2 and KANSL3 were found to bind ESC enhancers. There is evidence to suggest that NSL complex-associated MOF has specificity towards H4K5 and H4K8 (on recombinant nucleosomes; Cai et al 2010 PMID: 20018852) and H4K5 (in cultured fly cells; Feller et al 2015 PMID: 25578876) in addition to targeting H4K16. NSL complex-mediated acetylation promotes recruitment of BET domain-containing protein BRD4 in both flies and mammals (Gaub et al 2020 Nature Communications). NSL complex targets in mammals are diverse, including a combination of housekeeping and developmental genes.
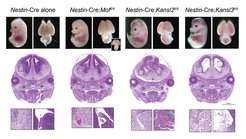
The lab has recently made a major push to investigate the tissue-specific functions of the NSL complex in mice. We have shown that mice with conditional deletion of Mof and Kansl2 via the hematopoiesis system-specific Vav1-iCre share phenotypic features, including decreased erythroid cells and their progenitors. On the other hand, Vav1-iCre-mediated loss of Kansl3 led to embryonic lethality (Pessoa Rodrigues et al 2020 Science Advances). We have also recently generated brain-specific knockout mice by specifically deleting Mof, Kansl2 and Kansl3 in embryonic brains using the Nestin-Cre. These mice die perinatally and suffer from brain hemorrhaging and neural defects. MRI analyses revealed a two-fold increase in the size of the lateral ventricles in Mof, Kansl2 and Kansl3 knockout brains at E14.5. Our analyses revealed that MOF or NSL complex loss in embryonic neurons leads to imbalance of long-chain fatty acids, which induce a TLR4-NFκB-driven inflammatory response in vascular pericytes, subsequently increasing vascular permeability and ultimately causing hemorrhaging (Sheikh et al 2020 Nature Cell Biology).
Histone acetylation-independent functions of MOF and MOF-associated complexes
In addition to its “canonical” role in the nucleus, as part of the NSL complex MOF also plays a key role in regulating transcription and respiration in mitochondria (Chatterjee et al. 2016 Cell). Initially by confocal microscopy, we detected a significant pool of MOF, as well as NSL complex members KANSL1, KANSL2 and KANSL3, in mitochondria. Gene expression analysis revealed that MOF regulates expression of mitochondrial genes involved in oxidative phosphorylation (OXPHOS) in aerobically respiring cells. Depletion of MOF or KANSL1 causes significant downregulation of mitochondrial DNA (mtDNA) transcription and subsequent translation, leading to impaired cellular respiration. Importantly, mitochondrial transcriptional and respiratory defects can be rescued by a mitochondrially tethered MOF, but not its catalytic mutant (E350Q), indicating that MOF’s enzymatic activity is important for mitochondrial function.
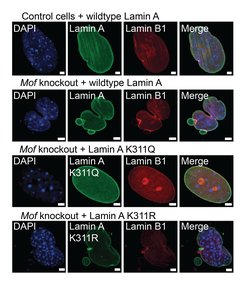
In addition to its roles in gene regulation, MOF has recently been shown to guard nuclear integrity through acetylation of a nuclear lamina protein. Using comprehensive acetylome screening we identified the structural protein Lamin A as a direct non-histone target of MOF. MOF associates with the NSL complex to acetylate Lamin A/C at lysine 311 (K311). Loss of MOF acetylation at Lamin A K311 has catastrophic consequences for the cell, including micronuclei formation, nuclear blebs, and chromothripsis (Karoutas et al. 2019 Nature Cell Biology). As part of this work we also identified multiple other non-histone targets of MOF acetylation which we are currently investigating.
Bioinformatic methods and technology development
When existing methodologies are unable to adequately answer our biological questions we are eager to devise new experimental or analysis approaches. Recently the lab has developed methods to accurately identify transcription start sites (Bhardwaj et al 2019 Nature Communications) and map the RNA-interacting domains within RNA-binding proteins (Panhale et al 2019 Nature Commmunications). Furthermore, former members of the lab have contributed to the deepTools2 suite (Ramírez et al 2016 Nucleic Acids Research) and snakePipes package (Bhardwaj et al 2019 Bioinformatics) developed by the bioinformatics facility headed by Dr. Thomas Manke.
Dr. Asifa Akhtar holds a patent for an optimised method to identify transcriptome-wide targets of RNA-binding proteins in vivo(International Patent Publication Number: WO/2017/013005; International Publication Date 26.01.2017; US Publication Number: US20180208966; EU Publication Number: EP3325621 A1.).





