Protocol Development
Standardization of workflows is key to robust, reproducible sequence data production.
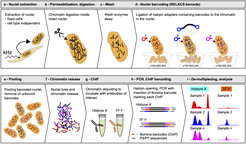
As partner of an international epigenome consortium (Stunnenberg et al., 2016) we developed standardized chromatin workflows to facilitate ChIP-Seq assays from a wide range of various primary tissues (Liver, Brain, Blood, Muscle) and different organisms (drosophila embryos, paramecium). To overcome the need of a cell type specific preparation method we introduced a method called Nuclei EXtraction by SONication (NEXSON) (Arrigoni et al., 2015). This method allows to extract nuclei from a wide range of formaldehyde fixed material. Isolated nuclei were processed with standard ChIP-Seq workflows. The method proved to be extremely versatile (histone modification and transcription factor maps) and sensitive (down to 10.000 cells per ChIP).

For standardized high-throughput ChIP-Seq a method called RELACS (restriction enzyme-based labeling of chromatin in situ) was introduced (Arrigoni et al., 2018). RELACS relies on standardized nuclei extraction from any source and employs chromatin cutting and barcoding within intact nuclei. Barcoded nuclei are pooled and processed within the same ChIP reaction, for maximal comparability and workload reduction. The innovative barcoding concept is particularly user-friendly and suitable for implementation to standardized large-scale clinical studies and scarce samples. Aiming to maximize universality and scalability, RELACS can generate ChIP-seq libraries for transcription factors and histone modifications from hundreds of samples within three days.
RELACS has been fully automated (Arrigoni et al., 2020) to support projects relying on high-throughput ChIP-Seq.